Optional exercises #1 :
If you have more time to spare and you are up for a challenge, take a look at the nucleosome. Right click here and save as pdb file. Open it from within spdbv. You might want to do some of the future exercises with the nucleosome in addition to the ATPases - thus save the pdb file, where you can find it again.
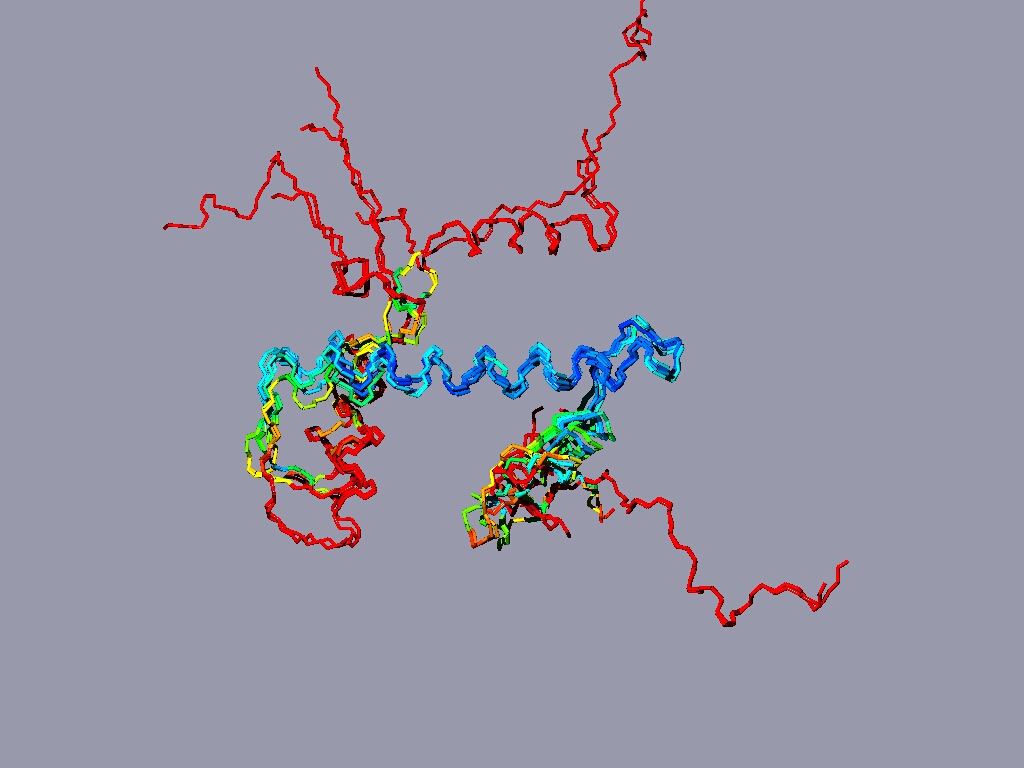
Align all the histones form the nucleosome to one reference histone and color in rmv:
The result might look something like this:
The picture shows a structure alignment of the 8 histones (2 each) that are part of the nucleosome. All the histones were colored regarding the match to H2A, except H2A, which was colored according to its match to H3. Coloring option RMS - shorter wavelengths - better match

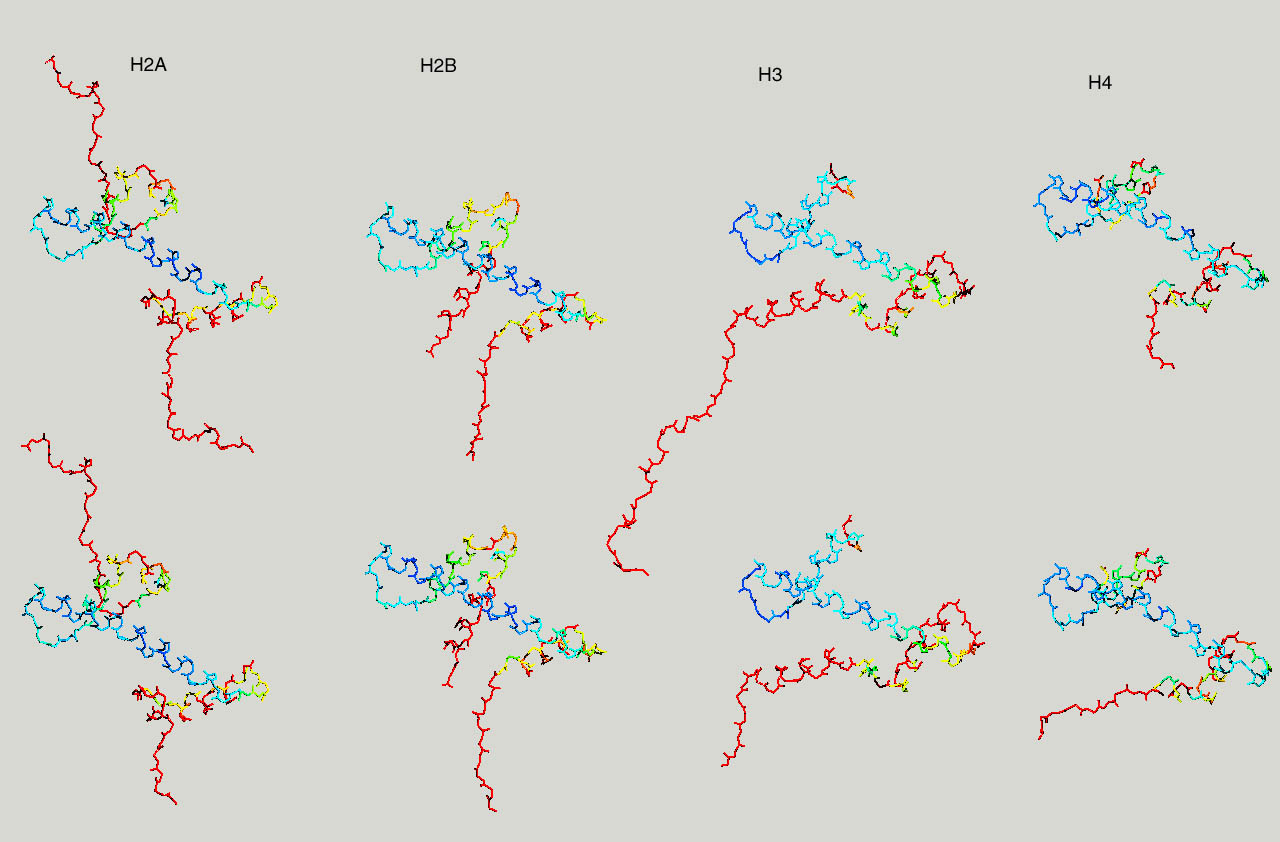
Below same as last figure, but histones are depicted side by side :
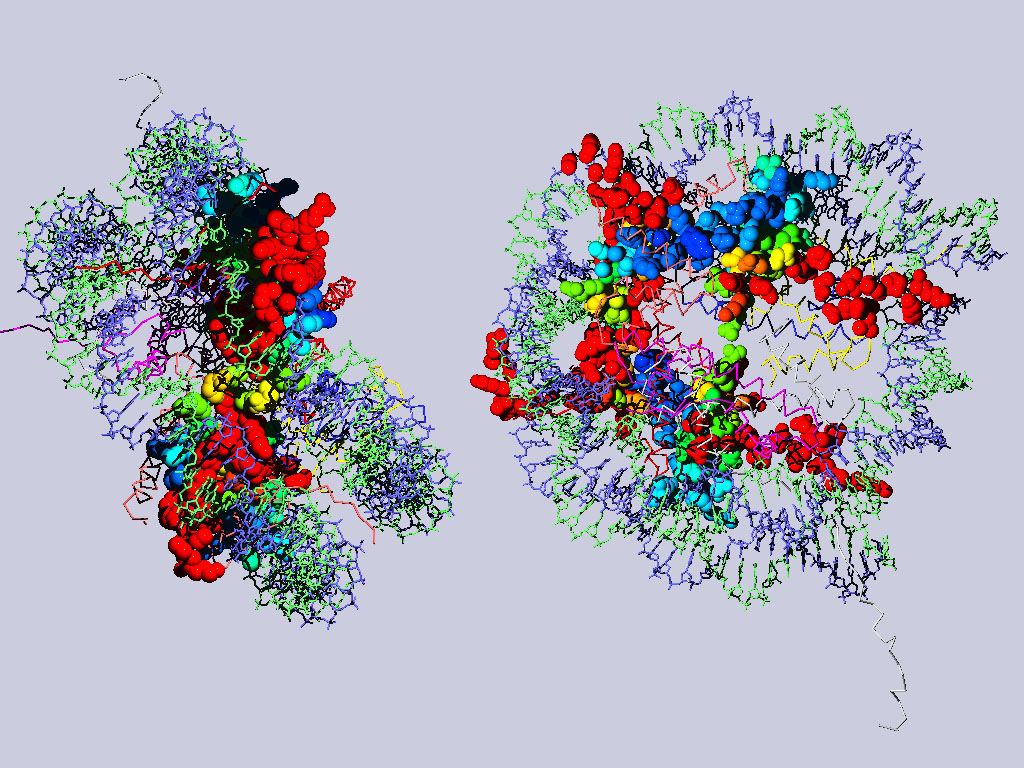
Below are two views of the complete nucleosome. Histones H2A are depicted as spacefilling balls and RMS colored regarding their match to H3. The rest of the molecule is colored according to chain.
Optional Exercise #2:
Load betaE, betaDP and betaTP into separate layers
Simplify the display and color all layers according to secondary structure.
Align betaE, betaDP and betaTP using magic fit followed by improve fit. (beta DP is a good reference layer).
Press control tab on the keyboard to quickly cycle through the aligned layers, which represent the changes the subunit undergoes in the catalytic cycle.
The same, but with the other subunits dragged along and aligned as well is here (saves as povray images and assembled into an animated gif).