CLASS 1. Introduction. Biological Databases.
Bioinformatcs Definition
Ch.1, Ch.2 (textbook);
Ch.1 [for background review], Ch.3 (supp. textbook);
Dobzhansky 1973 (Moodle) [optional, for historical interest]
- is an interdisciplinary area of research at the interface of computer science and biology
- specifically relates to quantitative analyses of data from biological micromolecules (DNA, RNA, protein)
- uses computers to store, retrieve and manipulate data (since most analyses are repetitive, mundane and complex)
- is a relatively new field that took off only after the first genome sequences became available, when it was recognized that efficient computation was needed to deal with hugh amount of sequence information
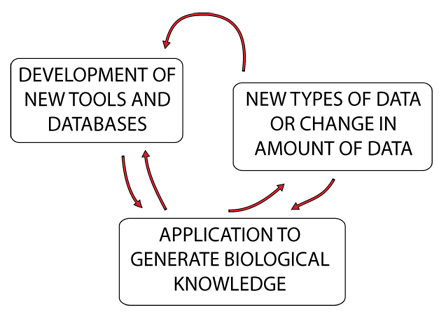
Bioinformatics Applications
In what research problems has bioinformatics shown to be of use?
- Applied Science: drug design, genetically-modified plants, forensic DNA analyses, genomics-based personalized medicine
- Molecular Dating (inferring dates of past evolutionary events and ancestral states of extinct organisms)
- Predicting existence of organisms with novel biochemistry
What does Molecular Evolution have to do with Bioinformatics?
Extrapolating from Central Dogma of Molecular Biology, the following chain is beleived to link primary (DNA) information of organism(s) with their phenotype and ecology.
DNA sequence 
transcription 
translation 
protein folding 
protein function (catalytic and other properties) 
organism(s) phenotype 
ecology  ... .
... .
The problem is that this chain is so complex, that currently it cannot be directly predicted given a DNA sequence. The solution: Use evolutionary context. Paraphrasing famous statement by Theodosius Dobzhansky, "Nothing in biologybioinformatics makes sense except in the light of evolution" (Dobzhansky, 1973, see homework reading). As we will see throughout the course, molecular evolution is indeed a foundation of bioinformatics.
Present day proteins evolved through substitutions and selection from ancestral proteins. As a corollary, related proteins have similar sequence AND similar structure AND similar function.
Experience shows that protein sequence space is so big that similar sequences do not arise through convergent evolution (at least if significant similarity is detectable through pairwise comparison). Here is back-of-the-envelope calculation:
There are 20 to the power of 600 = 4*10780 different proteins possible with lengths of 600 amino acids. For comparison the universe contains only about 1089 protons and has an age of about 5*1017 seconds or 5*1029 picoseconds. If every proton in the universe were a computer that explored one possible protein sequence per picosecond, we only would have explored 5*10118 sequences, i.e. a negligible fraction of the possible sequences with length 600 (one in about 10662).
Based on observation, the following principle emerges:
THE REVERSE IS NOT TRUE:
PROTEINS (or DOMAINS) WITH THE SAME OR SIMILAR FUNCTION DO NOT ALWAYS SHOW SIGNIFICANT SIMILARITY for one of
two reasons:
a) they evolved independently (e.g. different types of nucleotide binding sites)
b) they underwent so many substitution events that there is no readily detectable similarity remaining.
Moreover, due to reason (b), PROTEINS WITH SHARED ANCESTRY DO NOT ALWAYS SHOW SIGNIFICANT SIMILARITY.
Types of Information in Molecular Biology
Let's look at all databases at National Center for Biotechnology Information, using "all[filter]" as a query.
- DNA sequence [format]
- Protein Sequence [format]
- Structure (DNA, RNA, Protein)
- Whole Genomes
- Collective DNA from an environment (metagenomes)
- Gene Expression (Expressed Sequence Tags [ESTs], microarray experiments)
- Protein-Protein Interactions and Metabolic Pathways
- Phenotype experimental data
- Regulatory Networks
- Literature (PubMed)
- etc.
Biological Databases
A database is a searcheable archive to store and organize data (database records). Searching database with specific piece of information is called making a query.
- Primary (raw sequence data; e.g., GenBank, Protein Data Bank, Sequence Read Archive)
- Secondary (processed or curated data; e.g., RefSeq, UniProt, Protein Clusters)
- Specialized (e.g. specific molecule [Ribosomal Database Project], particular organism [Eukaryotic Genomes])
First sequence database was Atlas of Protein Sequence and Structure by Margaret Dayhoff (a book!). Went through five editions between 1965 and 1972. The first edition contained 93 one-sided pages [Source].
-
GenBank is a U.S.-based repository of primary sequence data. There are European (EMBL) and Japanese (DDJB) equivalents. These three databases exchange the data daily.
- Anatomy of a GenBank database record (a.k.a. GenBank Flat File Format). Note that the record contains not just sequence data, but a lot of auxilary information (annotations).
- GenBank Redundancy: GenBank contains many records multiple times. Examples of curated non-redundant databases: RefSeq, Swiss-Prot.
- Tracking Database Records: GeneInfo Identifier (GI numbers) vs. Accession Number. GI number is a unique numerical identifier associated with GenBank record. It is assigned sequentially to new records. If sequence in a record is updated, a new GI number is assigned. This allows tracking any changes to a record. Accession number is also a unique alphanumerical identifier in "accession.version" format. However, if any change happens to a sequence within the record, only "version" portion of the accession number changes. Accession numbers are used as unique identifiers by GenBank/EMBL/DDBJ. [more details]

NCBI (National Center for Biotechnology Information)
is a home for many public biological databases (see diagram below). All of the databases are interlinked,
and they all have common search and retrieval system - Entrez.

Fancier and more up-to-date diagram is here (requires Flash).
Want to be informed about new sequences/articles in your research area? Check out these services:
 |
PubCrawler |
| Swiss-Shop |
In short, the services allow a user to define queries that are stored in the user's profile. Using these queries the searches are regularly performed against the updates to the databases, and then the user is informed (by email alert) if there is anything new that match the queries. Great way to save time and stay up-to-date!
To construct a query, use Search Field Tags (new Search Field Tags for PubMed) and Boolean operators (AND, OR, NOT).
Explore Limits, Preview/Index, History, Clipboard and MyNCBI features of Entrez. (Since November 2009, PubMed has a re-designed interface and some of these features are located combined under "Advanced Search").
Databases at NCBI are interlinked and also provide links to outside resources (LinkOut).

