CLASS 10. Looking for distant homologs.
Ch.6 (textbook);
Sections 6.1, 6.2 (supp. textbook) [OPTIONAL]
Position-Specific Scoring Matrices [PSSM]
Regular substitution matrices (such as BLOSUM62 matrix shown above) are general scoring schemes. PSSMs can be viewed as extension of substitution scoring matrices, that take into account of probabilities of a certain amino acid to occur in a specific position in the protein. To generate a PSSM, sequences are required to be aligned.
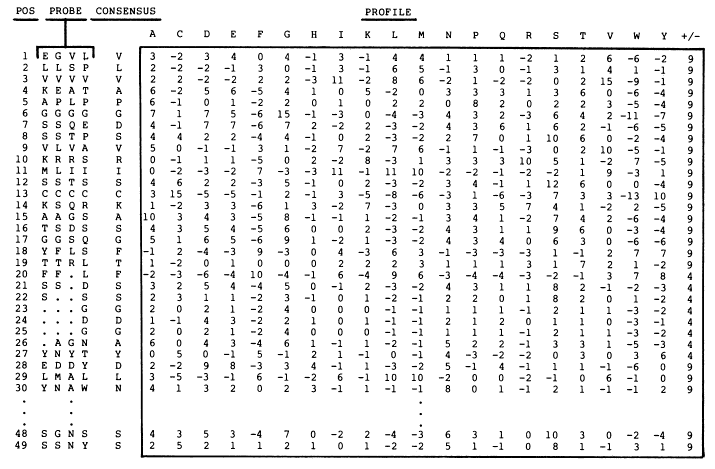
NCBI offers a PSSM viewer, where a PSSM matrix can be viewed color-coded, cross-linked to amino acid explorer and can be re-sorted by different features (e.g., examine PSSM-id 110296).
PSI-BLAST
Position-Specific Iterative (PSI) - BLAST is an enhanced BLAST program intended to help finding more distant homologs to a query sequence. A pre-requisite for it to work is presence of more closely related sequences in the database.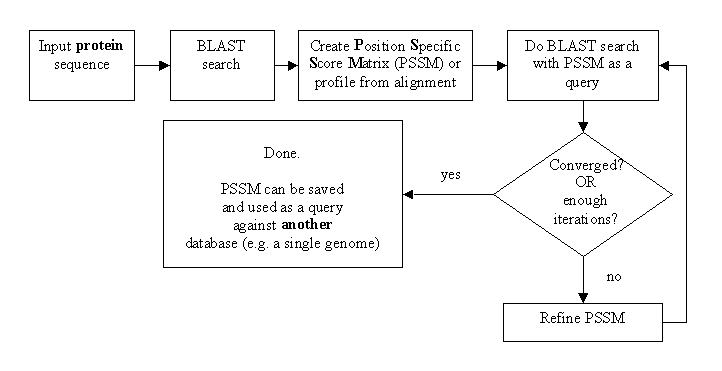
PSI-BLAST interactive tutorial shows step-by-step how PSI-BLAST works and how to use its web-based interface.
Constructed PSSMs can be saved and a database search can be made against the database of PSSMs. RPS-BLAST (Reverse Position Specific BLAST) program is used for this task. Example: Conserved Domains search.
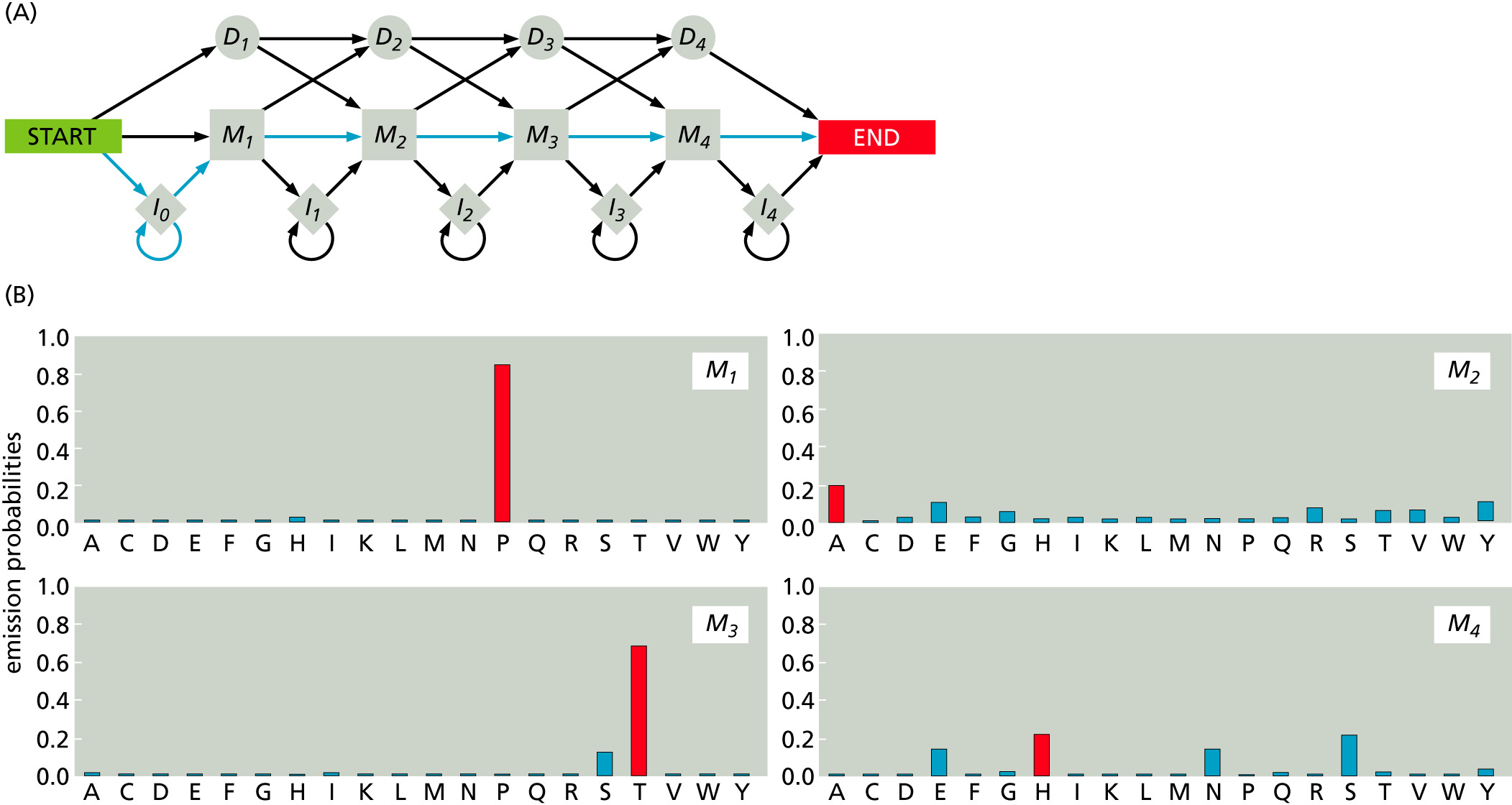
Hidden Markov Models
In general, a process has a Markov property if the probability of its future states depend only upon its present state and a fixed number of past states. Markov chain is a discrete random process with Markovian property. Markov chains often represented as directed graphs. For example, a DNA sequence can be shown as this Markov Model:
Components of a Markov Model:
- States (nodes on the graph)
- Transition probabilities (how one state changes to another state, shown as arrows on the graph)
- Emission probabilities
The i-th residue of a protein can be emitted from multiple alternative states. Since the residue itself does not carry information about which state it originated from, the model is referred as a "hidden" Markov model.
HMM applications in bioinformatics
- Modeling Protein Families (e.g., Superfamily DB)
- Characterization of Protein Domains (e.g., Pfam DB)
- Multiple Sequence Alignment (e.g., ProbCons program)
- Gene Finding (e.g., GENSCAN)