CLASS 12. Proteins in Bits and Pieces.
HOMEWORK READING:
Ch.7 (textbook);
Section 4.8-4.10, 6.6 and pp. 177-178 (supp. textbook)
Ch.7 (textbook);
Section 4.8-4.10, 6.6 and pp. 177-178 (supp. textbook)
Protein Motifs, Folds and Domains
A motif is a short conserved sequence pattern associated with distinct functions of a protein. Structurally, a motif is a simple combination of a few secondary structure elements. Example: helix-turn-helix motif, which is involved in DNA binding
A fingerprint is a group of conserved motifs used to characterise a protein family (note that motifs do not have to be adjacent to each other).
A protein domain is a well-defined region within a protein that either performs a specific function or constitutes a stable, compact structural unit within the protein that can be distinguished from all other parts (Wen-Hsiung Li, "Molecular Evolution", Sinauer, 1997). Protein can contain one or more domains. Domains usually consist of several motifs and folds. Example: human urokinase
Some reasons for attention to domains and motifs:
- A protein can have multiple functions (e.g., multidomain protein)
- Motifs can be short and divergent, and therefore can evade detection in a BLAST search
- Annotation of hypothetical proteins (which constitute ~30-40% sequenced proteins) is aided by identification of conserved elements. Interestingly, many of these hypothetical proteins turn out to belong to already known protein folds: see recent study here.
- PSSMs and HMMs
- Regular Expressions
- Sequence LOGOs
- Search a database of patterns with a query sequence (which may suggest functional activity of the query).
- Search a sequence database with a specific pattern (e.g., to examine how widespread a particular pattern is in different protein families).
- Define a pattern from a set of sequences.
Regular Expressions
Regular expression is a concise representation of a pattern (in our case a sequence family) as a string of characters.Basic Rules of a regular expression formation:
- Alternative amino acid residues are placed within square brackets
- Amino acids to be excluded are placed within curly braces
- Any residue is indicated by X
- Pattern/residue repetition is shown as one number in parentheses (n) or as a range (n,m)
- Each position is linked by a hyphen
- Pattern at amino terminal of sequence is denoted by <
- Pattern at carboxy-terminal of sequence is denoted by >
[LIVM]-x-[LIVM](2)-[HEA]-[TI]-x(1,3)-D-x-H-[GSA]-{P}-[LIVMF].
Query sequence is matched against the regular expression (or a database of regular expressions) using either exact matching or fuzzy matching (i.e., when some mismatches are allowed).
 is a curated sequence pattern database that initially was based solely on regular expressions (but now also uses profiles). Example of the pattern entry.
is a curated sequence pattern database that initially was based solely on regular expressions (but now also uses profiles). Example of the pattern entry.
Motif searching can also be done with Pattern Hit Initiated BLAST (PHI-BLAST). PHI-BLAST takes as input a query protein sequence and a regular expression and searches the specified database for other protein sequences that also contain the input pattern and have significant similarity to the query sequence in the vicinity. PHI-BLAST is located on the same page where BLASTP is (see prefilled example).
Searching for motifs and domains using PSSMs and HMM profiles
PRINTS - a curated database of protein fingerprints. E.g. ATP synthase subunit A fingerprint. is a database of domain profile HMMs produced from curated alignments (currently contains 866 entries).
is a database of domain profile HMMs produced from curated alignments (currently contains 866 entries).
Pfam is a database of alignments of protein domains that uses profile HMMs.
Conserved Domains Database (CDD) and InterPro are integrated databases that contain multiple motif/domain databases (including the ones described above). CD search is a part of every protein BLAST search. E.g, Pfam entry for ATP synthase subunit A in CDD.
Motif discovery without multiple sequence alignments
Alignments could contain errors or be unreliable because the sequences are either too divergent or vary dramatically in length. Therefore, it is also desirable to develop motif search methods that do not rely on pre-existing multiple sequence alignments. E.g., see MEME Suite or PRATT.Sequence Logo
An effective way to depict a motif is to use graphical representation, called sequence logo. Logos use the notion of sequence uncertainty, called Shannon entropy (from information theory):Hu=-Σfu,alog2fu,a
Iu=log220-Hu
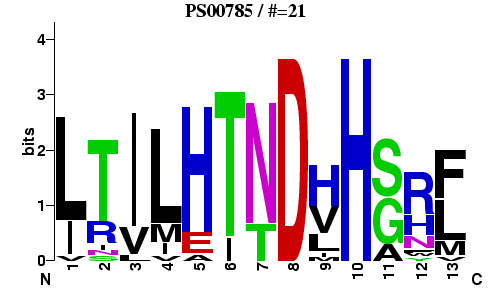
Fig. Sequence Logo for PROSITE entry PS00785.
21 true sequence matches were used to generate the logo.
21 true sequence matches were used to generate the logo.
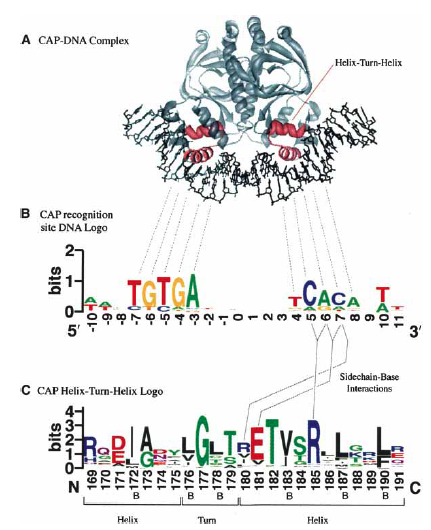
Fig. Helix-turn-helix motif containing CAP protein and sequence logos
for both DNA it binds to and HTH motif. [Source]
for both DNA it binds to and HTH motif. [Source]
More examples of sequence logos utility.
Web-based application to create sequence logos is called WebLOGO.
