CLASS 27. ASRV. Consensus Trees. Exploration of Tree Space. Parsimony. PHYLIP.
Ch. 10,11 (textbook);
Ch. 8 (supp. textbook);
Yang, 1996 (Moodle);
Among Site Rate Variation
Different regions of DNA or protein sequence may have different probabilities to undergo change. Two ways to deal with this problem mathematically:
- Allow only a fraction of sites to vary: X% of alignments sites are free to vary, whole remaining (100-X)% have probability zero to undergo change.
- Use Gamma distribution to describe how rates vary across sites:
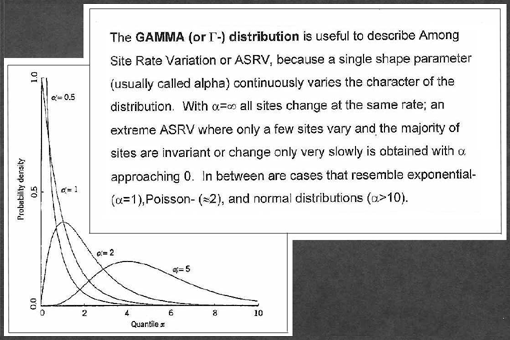
In phylogenetics, Gamma distribution is usually approximated by a discrete distribution (i.e. several rate categories are used, usually 4 or 8). If Gamma distribution is used in the model, it is designated by symbol Γ (e.g., 'HKY85+Γ'). For more information, see Z. Yang (1996) review (posted to Moodle).

ML methods with among site rate variation deal better with LBA artifact than parsimony and distance analyses (data)
Consensus Trees
- Strict Consensus Tree: only those clusters present in all of the trees will be present in the consensus tree.
- Majority Rule Consensus Tree: only those clusters present in the specified fraction (>0.50) of the trees will be present in the consensus tree (most often type of consensus tree used in bootstrap analyses.


Exploring Tree Space
Tree Space is vast and therefore if you have more than 20 sequences, it is not feasible to find an optimal tree through exhaustive search of tree space.
Heuristic Strategy:
- Start with a tree (often a NJ tree, can be a randomly chosen tree)
- Rearrange the tree and assess if it is better one (according to some "score" based on tree's relation to the data)
- Continue rearranging the tree, keeping only better rearrangements ("hill climbing")

How do trees get rearranged (illustration):
- Nearest Neighbor Interchange (NNI)
- Subtree Pruning and Regrafting (SPR)
- Tree Bisection and Reconnection (TBR)
Parsimony
Data for parsimony is individual sites in the alignment and number of changes for all sites are added up to get a score of a tree (unweighted parsimony). Most parsimonious tree is the tree that requires smallest number of evolutionary changes (has the smallest score).


Sites can be:
- informative (sites that discriminate alternative trees)
- uninformative (sites that do not discriminate alternative trees)
Concept of informative and uninformative sites applies only to parsimony analyses
Example:
sequence1 AATGC sequence2 AATCA sequence3 GATCA sequence4 GACCC
Objections to parsimony:
- Why should evolution be most parsimonious?
- Parsimony ignores branch lengths
- Parsimony is prone to long branch attraction (LBA).
PHYLIP

PHYLIP (the PHYLogeny Inference Package) is an open-source package of programs for inferring phylogenies (evolutionary trees), distributed and maintained by Joe Felsenstein and collaborators.
- PHYLIP is the oldest widely-distributed package. It has been in distribution since October 1980, and has over 28,000 registered users.
- Contains 35 command-line programs (modules)
- All programs are controlled through a text menu
- Output is written onto special files with names like "outfile" and "outtree".
- PHYLIP is very well documented: official and 3rd party manuals.